See also
A Jupyter notebook version of this tutorial can be downloaded here.
Binned acquisition
In this tutorial, we will demonstrate the sequencer-based acquisition binning procedure. The binning process is applied on the input path after real-time demodulation, (weighed) integration, IQ rotation and discretization. It allows storing both the integration and discretization results on the fly without intervention of the host PC in up to 131072 bins. It also allows the averaging of those bins on the fly (see section Sequencer Acquisition). We will show this by using a QRM and directly connecting outputs \(\text{O}^{[1-2]}\) to inputs \(\text{I}^{[1-2]}\), respectively. We will then use the QRM’s sequencers to sequence waveforms on the outputs and, simultaneously, acquire the resulting waveforms on the inputs.
To run this tutorial please make sure you have installed and enabled ipywidgets:
pip install ipywidgets
jupyter nbextension enable --py widgetsnbextension
Setup
First, we are going to import the required packages.
[1]:
# Import ipython widgets
import json
import math
import os
import ipywidgets as widgets
import matplotlib.pyplot
import numpy
# Set up the environment.
import scipy.signal
import ipywidgets as widgets
#from ipywidgets import fixed, interact, interact_manual, interactive
from IPython.display import display
from qblox_instruments import Cluster, PlugAndPlay
from qcodes import Instrument
from typing import List
Scan For Devices
We scan for the available devices connected via ethernet using the Plug & Play functionality of the Qblox Instruments package (see Plug & Play for more info).
[2]:
# Scan for available devices and display
with PlugAndPlay() as p:
# get info of all devices
device_list = p.list_devices()
names = {
dev_id: dev_info["description"]["name"] for dev_id, dev_info in device_list.items()
}
ip_addresses = {
dev_id: dev_info["identity"]["ip"] for dev_id, dev_info in device_list.items()
}
# create widget for names and ip addresses
connect = widgets.Dropdown(
options=[(names[dev_id] + " @" + ip_addresses[dev_id], dev_id)
for dev_id in device_list.keys()],
description="Select Device",
)
display(connect)
The following widget displays all the existing modules that are connected to your PC which includes the Pulsar modules as well as a Cluster. Select the device you want to run the notebook on.
Connect to Cluster
We now make a connection with the Cluster selected in the dropdown widget. We also define a function to find the modules we’re interested in. We select the readout and control module we want to use.
[ ]:
# Connect to device
dev_id = connect.value
# Close the chosen QCodes instrument as to prevent name clash.
try:
Instrument.find_instrument(names[dev_id]).close()
except KeyError:
pass
cluster = Cluster(name=names[dev_id], identifier=ip_addresses[dev_id], debug=True) # TODO remove debug
print(f"{connect.label} connected")
print(cluster.get_system_state())
[ ]:
def select_module_widget(device, select_all=False, select_qrm_type: bool=True, select_rf_type: bool=False):
"""Create a widget to select modules of a certain type
default is to show only QRM baseband
Args:
devices : Cluster we are currently using
select_all (bool): ignore filters and show all modules
select_qrm_type (bool): filter QRM/QCM
select_rf_type (bool): filter RF/baseband
"""
options = [[None, None]]
for module in device.modules:
if module.present():
if select_all or (module.is_qrm_type == select_qrm_type and module.is_rf_type == select_rf_type):
options.append(
[
f"{device.name} "
f"{module.short_name} "
f"({module.module_type}{'_RF' if module.is_rf_type else ''})",
module,
]
)
widget = widgets.Dropdown(options=options)
display(widget)
return widget
[ ]:
print("Select the readout module from the available modules:")
select_readout_module = select_module_widget(cluster, select_qrm_type=True, select_rf_type=False)
[ ]:
readout_module = select_readout_module.value
print(f"{readout_module} connected")
Reset the Cluster
We reset the Cluster to enter a well-defined state. Note that resetting will clear all stored parameters, so resetting between experiments is usually not desirable.
[ ]:
cluster.reset()
print(cluster.get_system_state())
Generate waveforms and weights
Next, we need to create the waveforms used by the sequence for playback on the outputs as well as weights used by the sequence for weighed integrations. To keep it straightforward, we use the DC offset from the sequencers as our waveform and define waveform weights in the cell below.
[4]:
# Waveform and weight parameters
waveform_weight_length = 600 # nanoseconds
# These will be used as weights in the "Weighed acquisition" section
waveforms_weights = {
"gaussian": {
"data": scipy.signal.gaussian(
waveform_weight_length, std=0.12 * waveform_weight_length
).tolist(),
"index": 0,
},
"sine": {
"data": [
math.sin((2 * math.pi / waveform_weight_length) * i)
for i in range(0, waveform_weight_length)
],
"index": 1,
},
"block": {"data": [1.0 for _ in range(0, waveform_weight_length)], "index": 2},
}
Specify acquisitions
We also need to specify the acquisitions so that the instrument can allocate the required memory for its acquisition list. In this case we will create 4 acquisition specifications that each create multiple bins.
[5]:
# Acquisitions
acquisitions = {
"non_weighed": {"num_bins": 10, "index": 0},
"weighed": {"num_bins": 10, "index": 1},
"large": {"num_bins": 131072, "index": 2},
"avg": {"num_bins": 10, "index": 3},
}
Create Q1ASM program
Now that we have the waveform and acquisition specifications for the sequence, we need a simple Q1ASM program that sequences the waveforms and triggers the acquisitions. In this case we will simply trigger 10 non-weighed acquisitions and store each acquisition in a separate bin.
[6]:
# Sequence program.
seq_prog = """
move 0,R0 #Loop iterator.
nop
loop: acquire 0,R0,1200 #Acquire bins and store them in "non_weighed" acquisition.
add R0,1,R0 #Increment iterator
nop #Wait a cycle for R0 to be available.
jlt R0,10,@loop #Run until number of iterations is done.
stop #Stop.
"""
Upload sequence
Now that we have the waveform, weights and acquisition specifications and Q1ASM program, we can combine them in a sequence stored in a JSON file.
[7]:
# Add sequence program, waveforms, weights and acquisitions to single dictionary and write to JSON file.
sequence = {
"waveforms": waveforms_weights,
"weights": waveforms_weights,
"acquisitions": acquisitions,
"program": seq_prog,
}
with open("sequence.json", "w", encoding="utf-8") as file:
json.dump(sequence, file, indent=4)
file.close()
Let’s write the JSON file to the instruments. We will use sequencer 0, which will drive outputs \(\text{O}^{[1-2]}\) and acquire on inputs \(\text{I}^{[1-2]}\).
[8]:
# Upload sequence.
readout_module .sequencer0.sequence("sequence.json")
Play sequence
The sequence has been uploaded to the instrument. Now we need to configure the sequencers. To keep it simple we will set a DC signal on the outputs of the instrument by enabling the sequencer offsets and disabling the modulation. These DC signals will then be acquired through the inputs. As such, we will also disable the demodulation on the input path. Furthermore, since we are running non-weighed integrations, we need to specify the integration length. This integration length will be used for every non-weighed integration moving forward. We will also put the integration result acquisition rotation to 0 degrees and acquisition threshold to 0.
[9]:
# Configure scope mode
readout_module.scope_acq_sequencer_select(0)
readout_module.scope_acq_trigger_mode_path0("sequencer")
readout_module.scope_acq_trigger_mode_path1("sequencer")
# Configure the sequencer
readout_module.sequencer0.offset_awg_path0(0.5)
readout_module.sequencer0.offset_awg_path1(0.5)
readout_module.sequencer0.mod_en_awg(False)
readout_module.sequencer0.demod_en_acq(False)
readout_module.sequencer0.integration_length_acq(1000)
readout_module.sequencer0.thresholded_acq_rotation(0)
readout_module.sequencer0.thresholded_acq_threshold(0)
# Map sequencer to specific outputs (but first disable all sequencer connections)
for sequencer in readout_module.sequencers:
for out in range(0, 2):
sequencer.set("channel_map_path{}_out{}_en".format(out % 2, out), False)
readout_module.sequencer0.channel_map_path0_out0_en(True)
readout_module.sequencer0.channel_map_path1_out1_en(True)
Now let’s start the sequence.
[10]:
# Arm and start sequencer.
readout_module.arm_sequencer(0)
readout_module.start_sequencer()
# Print status of sequencer.
print(readout_module.get_sequencer_state(0, 1))
Status:
Status: STOPPED, Flags: ACQ_SCOPE_DONE_PATH_0, ACQ_SCOPE_DONE_PATH_1, ACQ_BINNING_DONE
Retrieve acquisition
Next, we will have a quick look at the input signal so that we can compare it to the integration results. Since we are integrating over a DC signal we are expecting the integration results to be roughly equal to the average DC value.
[11]:
# Wait for the sequencer to stop with a timeout period of one minute.
readout_module.get_acquisition_state(0, 1)
# Move acquisition data from temporary memory to acquisition list.
readout_module.store_scope_acquisition(0, "non_weighed")
# Get acquisition list from instrument.
non_weighed_acq = readout_module.get_acquisitions(0)["non_weighed"]
# Plot acquired signal on both inputs.
fig, ax = matplotlib.pyplot.subplots(1, 1, figsize=(15, 15 / 2 / 1.61))
ax.plot(non_weighed_acq["acquisition"]["scope"]["path0"]["data"][0:1000])
ax.plot(non_weighed_acq["acquisition"]["scope"]["path1"]["data"][0:1000])
ax.set_xlabel("Time (ns)")
ax.set_ylabel("Relative amplitude")
matplotlib.pyplot.show()
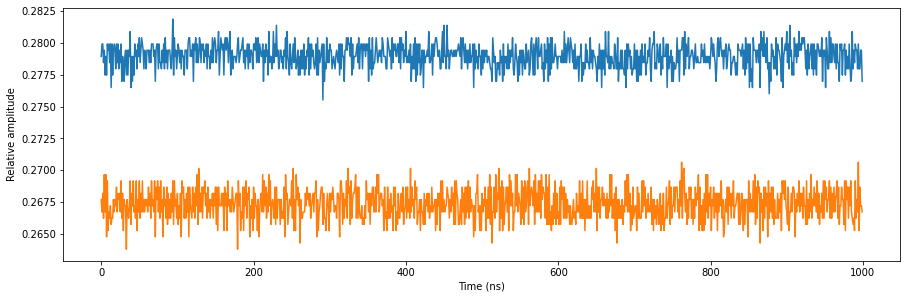
To check if the integration results match what we expect, we need to divide the integration results by the integration length which was set through the corresponding QCoDeS parameter. Note that the ‘valid’ key of the dictionary indicates if the bin was actually set during the sequence.
[12]:
int_len = readout_module.sequencer0.integration_length_acq()
bins = non_weighed_acq["acquisition"]["bins"]
bins["integration"]["path0"] = [(val / int_len) for val in bins["integration"]["path0"]]
bins["integration"]["path1"] = [(val / int_len) for val in bins["integration"]["path1"]]
bins
[12]:
{'integration': {'path0': [0.27915486077186125,
0.2791612115290669,
0.27915192965315094,
0.27901367855398146,
0.2791001465559355,
0.2791299462628236,
0.2789799706888129,
0.2790600879335613,
0.279057156814851,
0.27897557401074746],
'path1': [0.2676213971665853,
0.26770151441133366,
0.2676560820713239,
0.26742061553492913,
0.2675017098192477,
0.26750122129946263,
0.2674181729360039,
0.26754714215925746,
0.26734391792867607,
0.26737909135319976]},
'threshold': [1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0],
'avg_cnt': [1, 1, 1, 1, 1, 1, 1, 1, 1, 1]}
Weighed acquisition
In the following, we look into weighed integrations. To do this, we will need to modify the sequence program slightly and reupload it. We will be using a gaussian weight to integrate over input path 0 and a sine weight to integrate over input path 1. The integration length of a weighed integration is determined by the weight length.
[13]:
# Sequence program.
seq_prog = """
move 0,R0 #Loop iterator.
move 0,R1 #Weight for path 0.
move 1,R2 #Weight for path 1.
nop
loop: acquire_weighed 1,R0,R1,R2,1200 #Acquire bins and store them in "weighed" acquisition.
add R0,1,R0 #Increment iterator
nop #Wait a cycle for R0 to be available.
jlt R0,10,@loop #Run until number of iterations is done.
stop #Stop.
"""
[14]:
# Add sequence program, waveforms, weights and acquisitions to single dictionary and write to JSON file.
sequence = {
"waveforms": waveforms_weights,
"weights": waveforms_weights,
"acquisitions": acquisitions,
"program": seq_prog,
}
with open("sequence.json", "w", encoding="utf-8") as file:
json.dump(sequence, file, indent=4)
file.close()
[15]:
# Upload sequence.
readout_module.sequencer0.sequence("sequence.json")
Let’s start the sequence and retrieve the results.
[16]:
# Arm and start sequencer.
readout_module.arm_sequencer(0)
readout_module.start_sequencer()
# Print status of sequencer.
print(readout_module.get_sequencer_state(0, 1))
Status:
Status: STOPPED, Flags: ACQ_SCOPE_DONE_PATH_0, ACQ_SCOPE_DONE_PATH_1, ACQ_BINNING_DONE
[17]:
# Wait for the sequencer to stop with a timeout period of one minute.
readout_module.get_acquisition_state(0, 1)
# Get acquisition list from instrument.
weighed_acq = readout_module.get_acquisitions(0)["weighed"]
To check if the integration results match what we expect we need to divide the integration results by the integration length again. In this case the integration length is determined by the length of the weights.
[18]:
int_len = waveform_weight_length
bins = weighed_acq["acquisition"]["bins"]
bins["integration"]["path0"] = [(val / int_len) for val in bins["integration"]["path0"]]
bins["integration"]["path1"] = [(val / int_len) for val in bins["integration"]["path1"]]
bins
[18]:
{'integration': {'path0': [0.0838375615631583,
0.08379245402794945,
0.0838248975170451,
0.08380893490833831,
0.08373342157077765,
0.0837548154917014,
0.08381608371642879,
0.08383202245997753,
0.08376547844431308,
0.08379022621544653],
'path1': [-5.6059653985437636e-05,
-3.018027660831874e-05,
3.383960084091271e-05,
6.327051148681764e-06,
-1.1236511909867344e-06,
1.715626436135604e-05,
-0.00011699814177924457,
-5.624102918653143e-05,
-3.3345592069512524e-05,
-0.00010118747457863478]},
'threshold': [1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0],
'avg_cnt': [1, 1, 1, 1, 1, 1, 1, 1, 1, 1]}
Large number of bins
The QRM supports up to 131072 bins. To show that we need to change the program slightly. We will use the non-weighed acquisition program, however, we will now loop over the maximum number of acquisitions while storing each result in a separate bin.
[19]:
# Sequence program.
seq_prog = """
move 0,R0 #Loop iterator.
nop
loop: acquire 2,R0,1200 #Acquire bins and store them in "large" acquisition.
add R0,1,R0 #Increment iterator
nop #Wait a cycle for R0 to be available.
jlt R0,131072,@loop #Run until number of iterations is done.
stop #Stop.
"""
[20]:
# Add sequence program, waveforms, weights and acquisitions to single dictionary and write to JSON file.
sequence = {
"waveforms": waveforms_weights,
"weights": waveforms_weights,
"acquisitions": acquisitions,
"program": seq_prog,
}
with open("sequence.json", "w", encoding="utf-8") as file:
json.dump(sequence, file, indent=4)
file.close()
[21]:
# Upload sequence.
readout_module.sequencer0.sequence("sequence.json")
Let’s start the sequence and retrieve the results.
[22]:
# Arm and start sequencer.
readout_module.arm_sequencer(0)
readout_module.start_sequencer()
# Print status of sequencer.
print(readout_module.get_sequencer_state(0, 1))
Status:
Status: STOPPED, Flags: ACQ_SCOPE_DONE_PATH_0, ACQ_SCOPE_DONE_PATH_1, ACQ_BINNING_DONE
[23]:
# Wait for the sequencer to stop with a timeout period of one minute.
readout_module.get_acquisition_state(0, 1)
# Get acquisition list from instrument.
large_acq = readout_module.get_acquisitions(0)["large"]
Since the number of bins is now to large to simply print, we will check the number of bins and we will check the bins for NaN values which indicate that a bin is not written.
[24]:
int_len = readout_module.sequencer0.integration_length_acq()
bins = large_acq["acquisition"]["bins"]
bins["integration"]["path0"] = [(val / int_len) for val in bins["integration"]["path0"]]
bins["integration"]["path1"] = [(val / int_len) for val in bins["integration"]["path1"]]
print("Number of bins: {}".format(len(bins["avg_cnt"])))
for it, val in enumerate(bins["integration"]["path0"]):
if math.isnan(val):
Exception("NaN found at index {}.".format(it))
for it, val in enumerate(bins["integration"]["path0"]):
if math.isnan(val):
Exception("NaN found at index {}.".format(it))
for it, val in enumerate(bins["integration"]["path0"]):
if math.isnan(val):
Exception("NaN found at index {}.".format(it))
print("All values are valid.")
Number of bins: 131072
All values are valid.
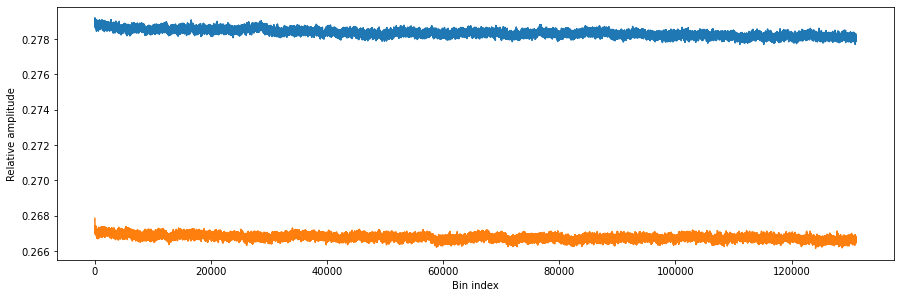
We will also plot the integration results in every bin to visualize the contents.
[25]:
# Plot bins
fig, ax = matplotlib.pyplot.subplots(1, 1, figsize=(15, 15 / 2 / 1.61))
ax.plot(bins["integration"]["path0"])
ax.plot(bins["integration"]["path1"])
ax.set_xlabel("Bin index")
ax.set_ylabel("Relative amplitude")
matplotlib.pyplot.show()
Averaging
As you may have noticed, the acquisition results also contain an average counter. This average counter reflects the number of times a bin has been averaged during the sequence. Each time the sequencer writes to the same bin the results are automatically accumulated and the average counter is increased. Upon retrieval of the acquisition results, each result is divided by the average counter and therefore automatically averaged. To show this we will change the sequence one last time. This time we will average 10 bins a 1000 times each.
[26]:
# Sequence program.
seq_prog = """
move 0,R1 #Average iterator.
avg: move 0,R0 #Bin iterator.
nop
loop: acquire 3,R0,1200 #Acquire bins and store them in "avg" acquisition.
add R0,1,R0 #Increment bin iterator
nop #Wait a cycle for R0 to be available.
jlt R0,10,@loop #Run until number of avg iterations is done.
add R1,1,R1 #Increment avg iterator
nop #Wait a cycle for R1 to be available.
jlt R1,1000,@avg #Run until number of average iterations is done.
stop #Stop.
"""
[27]:
# Add sequence program, waveforms, weights and acquisitions to single dictionary and write to JSON file.
sequence = {
"waveforms": waveforms_weights,
"weights": waveforms_weights,
"acquisitions": acquisitions,
"program": seq_prog,
}
with open("sequence.json", "w", encoding="utf-8") as file:
json.dump(sequence, file, indent=4)
file.close()
[28]:
# Upload sequence.
readout_module.sequencer0.sequence("sequence.json")
Let’s start the sequence and retrieve the results.
[29]:
# Arm and start sequencer.
readout_module.arm_sequencer(0)
readout_module.start_sequencer()
# Print status of sequencer.
print(readout_module.get_sequencer_state(0, 1))
Status:
Status: STOPPED, Flags: ACQ_SCOPE_DONE_PATH_0, ACQ_SCOPE_DONE_PATH_1, ACQ_BINNING_DONE
Note that the average count of each bin is now set to a 1000.
[30]:
# Wait for the sequencer to stop with a timeout period of one minute.
readout_module.get_acquisition_state(0, 1)
# Get acquisition list from instrument.
avg_acq = readout_module.get_acquisitions(0)["avg"]
[31]:
int_len = readout_module.sequencer0.integration_length_acq()
bins = avg_acq["acquisition"]["bins"]
bins["integration"]["path0"] = [(val / int_len) for val in bins["integration"]["path0"]]
bins["integration"]["path1"] = [(val / int_len) for val in bins["integration"]["path1"]]
bins
[31]:
{'integration': {'path0': [0.2783555486077186,
0.2783564631167562,
0.2783504118221788,
0.2783517176355642,
0.27834840449438203,
0.27835067464582314,
0.2783542623351246,
0.2783474118221788,
0.2783547987298486,
0.2783543351245726],
'path1': [0.26682932535417686,
0.2668245642403517,
0.26682411724474836,
0.2668218031265267,
0.2668268925256473,
0.26682839179286766,
0.2668272222765022,
0.2668283277967758,
0.2668281695163654,
0.2668296311675623]},
'threshold': [1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0],
'avg_cnt': [1000, 1000, 1000, 1000, 1000, 1000, 1000, 1000, 1000, 1000]}
Stop
Finally, let’s stop the sequencers if they haven’t already and close the instrument connection. One can also display a detailed snapshot containing the instrument parameters before closing the connection by uncommenting the corresponding lines.
[ ]:
# Stop sequencer.
readout_module.stop_sequencer()
# Print status of sequencer.
print(readout_module.get_sequencer_state(0))
print()
# Uncomment the following to print an overview of the instrument parameters.
# Print an overview of the instrument parameters.
# print("Snapshot:")
# readout_module.print_readable_snapshot(update=True)
# Close the instrument connection.
Cluster.close_all()